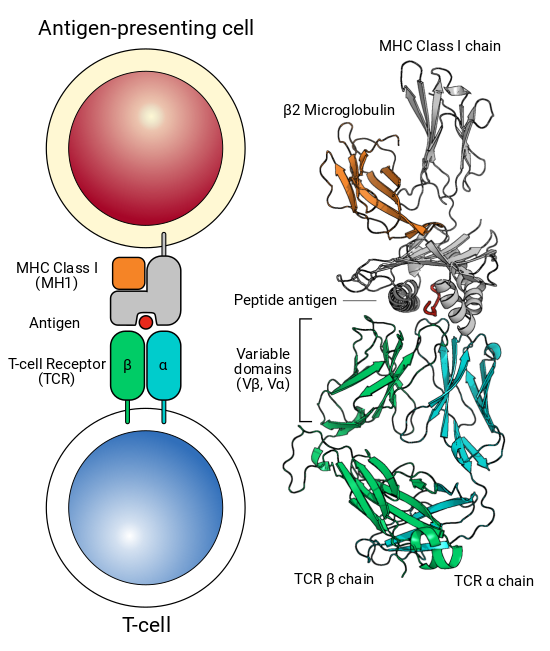
The following fields can be found in each summary file:
| pdb | The PDB code of the entry |
| Bchain | The name of the β chain, if present; else "NA" |
| Achain | The name of the α chain, if present; else "NA" |
| Dchain | The name of the γ chain, if present; else "NA" |
| Gchain | The name of the δ chain, if present; else "NA" |
| TCRtype | Detected TCR type; either "abTCR" (for αβTCR) or "gdTCR" (for γδTCR) or "NA" for single-domain cases. |
| model | Model number; defaults to 0 for structures solved by X-ray crystallography. |
| antigen_chain | The name of the antigen (i.e. non-TCR/non-MHC) chain, if present; else "NA" |
| antigen_type | Type of antigen. Either "peptide", "protein", "hatpen" or "carbohydrate". |
| antigen_name | Name of the antigen. |
| antigen_het_name | The HETATM name of the antigen. |
| mhc_type | Type of MHC unit. Either MH1, MH2, CD1, MR1 if present; else "NA". |
| mhc_chain1 | This is either MH1, CD1 or MR1 (for MHC class I, CD1 or MR1 proteins) or GA (for MHC class II), if present; else "NA" |
| mhc_chain2 | This is either B2M (for MHC class I, CD1 or MR1 proteins) or GB (for MHC class II), if present; else "NA" |
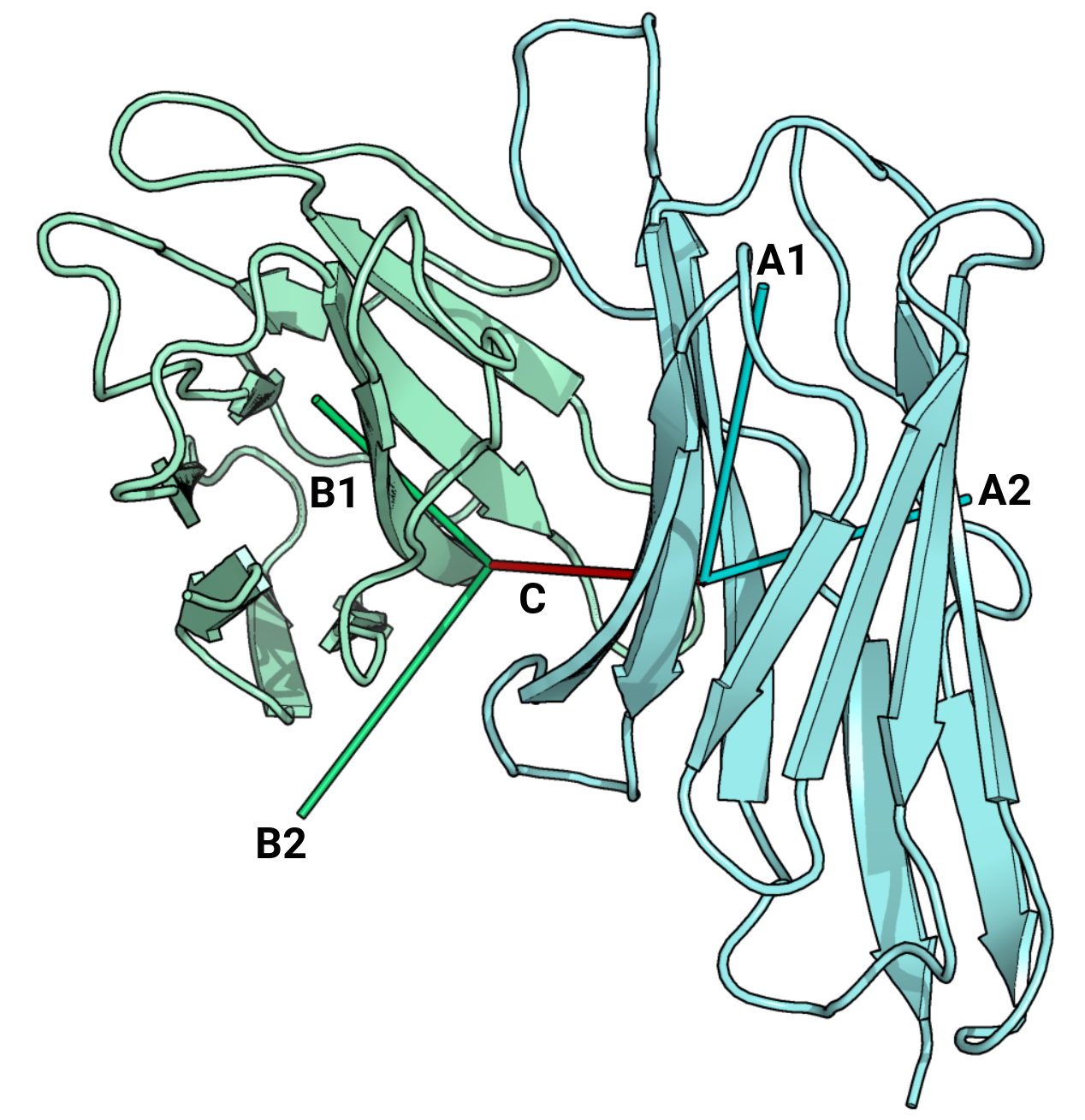
| docking_angle | Docking angle between the TCR and MHC |
| beta_subgroup | The IMGT subgroup for the β chain |
| alpha_subgroup | The IMGT subgroup for the α chain |
| delta_subgroup | The IMGT subgroup for the δ chain |
| gamma_subgroup | The IMGT subgroup for the γ chain |
| short_header | A short description of the structure from the PDB entry. |
| date | Deposition date of the structure. |
| compound | Name of the structure from the PDB. |
| beta_organism | The organism of the structure's β chain according to the PDB. |
| alpha_organism | The organism of the structure's α chain according to the PDB. |
| delta_organism | The organism of the structure's δ chain according to the PDB. |
| gamma_organism | The organism of the structure's γ chain according to the PDB. |
| antigen_organism | The organism of the structure's antigen chain(s) according to the PDB. |
| mhc_chain1_organism | The organism of the structure's first MHC chain (see mhc_chain1) according to the PDB. |
| mhc_chain2_organism | The organism of the structure's second MHC chain (see mhc_chain2) according to the PDB. |
| authors | Name(s) of author(s) that deposited the structure. |
| resolution | Resolution of the structure in Å. |
| method | Method used to solve the structure. |
| r_free | R-free value of the structure. |
| r_factor | R-factor value of the structure. |
| affinity | Affinity of the TCR, if present; else "NA" |
| affinity_method | Method used to measure the affinity. |
| affinity_temperature | Temperature of the conditions during affinity measurement. "NA" if unavailable. |
| affinity_pmid | Pubmed ID for publication with affinity measurement. |
| engineered | Indicates whether or not the structure is engineered. |