About OTS
Sequencing of TCR repertoires is known to be a hard task due to the need to physically link or label natively paired alpha and beta chains. With the advent of 10xGenomics sequencing, full-length natively paired Alpha/Beta sequences can now be obtained, although at the expense of the T cell throughput number in comparison to unpaired Illumina sequencing.
The Observed TCR Space (OTS) database now provides access to annotated paired sequences from 10xGenomics studies. To date, OTS collates over 1.6M non redundant paired sequences from 50 different studies. The data is available for download or you can filter the sequences with respect to certain metadata parameters using our search form. To download the data go to the Search page.
Paired sequences in OTS can be filtered according to attributes such as species, disease, treatment etc. The fields are non-exclusive, meaning that the user could choose a combination of fields that does not exist in our database.
Metadata
Similarly to the unpaired version of OAS, all datasets are organized into studies, that are in turn subdivided into data-units. A single data-unit is a set of sequences uniquely identified by its metadata. The range of meta-parameters are:
- Age Information on age of the human T cell donors.
- Disease Indicates the disease state of the donor or tissue sample at the time of T cell extraction.
- CancerType If disease is cancer, this indicates the type of malignacy.
- Treatment Indicates what type of treatment was administered to the donor.
- TType Top level descriptor of the sorted T cells or population from which the T cells were isolated.
- TSubtype Further sorting method or antigen enrichment strategy performed to obtain the T cell population.
- Species Organism of the T cell donor cells.
- Strain If mouse, indicates the strain or genotype.
- Author First author and the publication year.
- Sequences Number of sequences that passed quality filtering steps.
- TSource Which organ/tissue the T cells were extracted from.
- Subject Indicates whether the T cells can be tracked back to a particular individual.
- Longitudinal If the study is conducted over a period of time, indicates the particular timepoint when T cells were sourced.
Linking alpha and beta chains
In the ideal case scenario, 10xGenomics sequencing would yield 1-to-1 alpha/beta chain pairings for each interrogated T cell. However, in many cases more than one alpha and/or beta chain sequences harbour identical 10xGenomics cell barcodes. Linking such alpha and beta chain sequences can lead to combinatorial inflation of the real sequence number and incorrect estimation of the repertoire diversity - see example relating to antibody heavy and light chains (Figure 2).
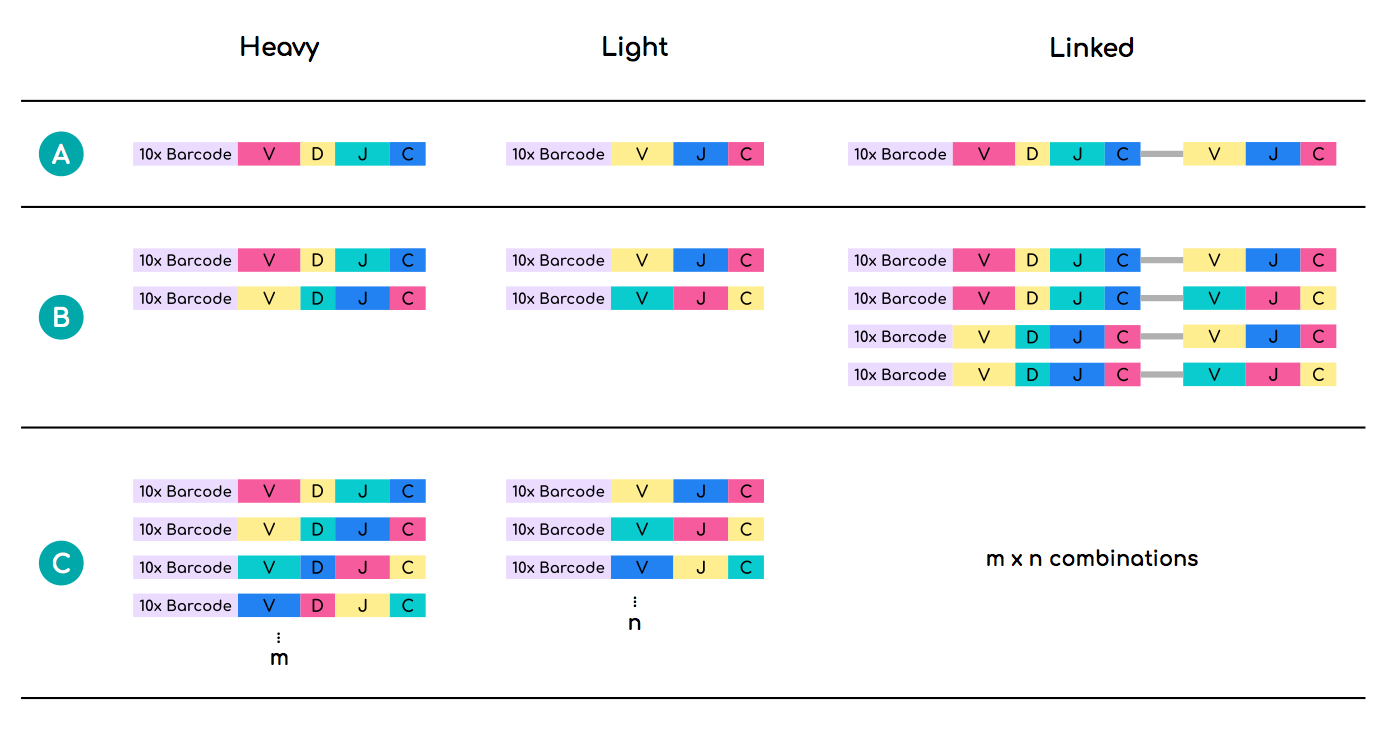
One solution is to filter out sequences whose 10xGenomics barcodes are shared between more than one unique alpha and beta V(D)J recombination events. This step has already been performed for each data unit in OTS.
Contact
If you would like to contact us or to submit your study to OTS, please drop an email to opig@stats.ox.ac.uk.
