About SAbDab
SAbDab is a database of antibody structures that updates on a weekly basis. Each structure is annotated with a number of properties including experimental details, antibody nomenclature (e.g. heavy-light pairings), curated affinity data and sequence annotations.
Use the database to:
- Inspect individual structures
- Create and download datasets for analysis
- Search the database for structures with similar sequences to your query
- Monitor the known structural repetoire of antibodies
SAbDab has been built by the Oxford Protein Informatics Group (OPIG) under an open-innovation agreement.
If you use our tools, please cite our paper: Dunbar, J., Krawczyk, K. et al (2014). Nucleic Acids Res. 42. D1140-D1146 [link]
Terminology
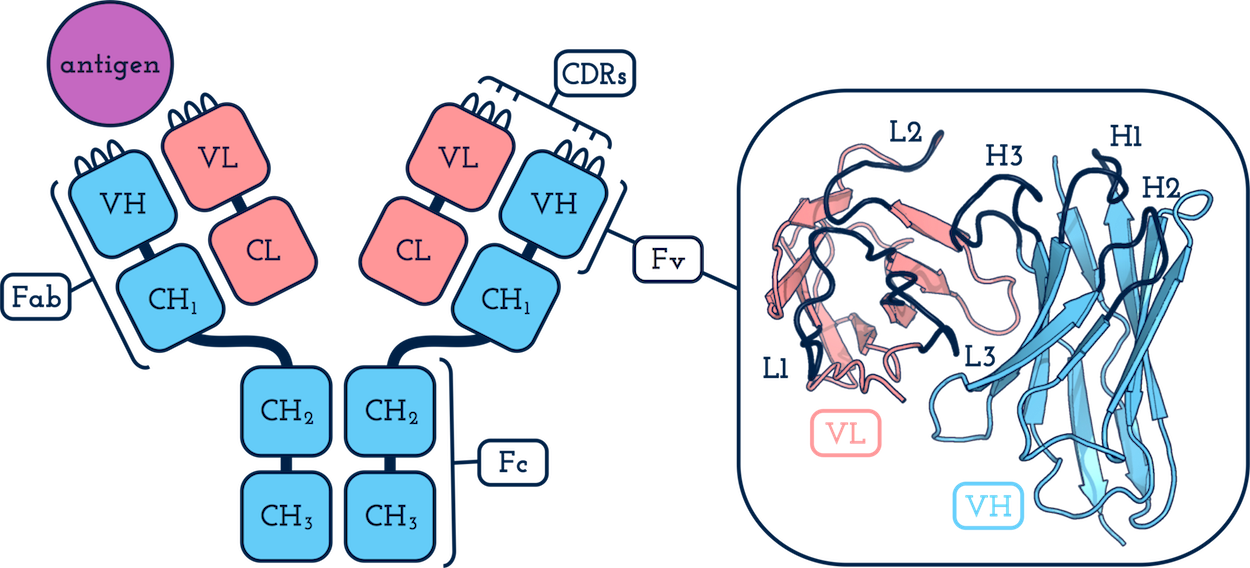
What is:
- an antibody? - an immunoprotein responsible for specifically recognising and binding to potentially pathogenic molecules.
- an antigen? - the molecule that the antibody targets.
- a heavy chain? - the longer antibody chain. This folds to form one variable domain (VH) and three or more constant domains (CH1, CH2 and CH3).
- a light chain? - the shorter antibody chain. This folds to form one variable domain (VL) and one constant domain (CL1).
- a variable domain (VH or VL)? - the variable part of the antibody.
- a CDR? - a Complementarity Determining Region. These are generally characterised by six hypervariable loops: three on VH (H1, H2 and H3) and three on VL (L1, L2 and L3). Diversity in the sequence and structure of the CDRs are the main determinants of antigen specificity and affinity.
- the framework region? - the collective name for the residues in VH or VL that are not CDRs.
- an Fv? - a variable fragment. The collective name for a paired VH and VL.
- an Fab?- an antigen binding fragment. The same as an Fv but also including the first constant domains of each chain, CH1 and CL1. Typically an antibody will have two Fab arms (see above). Most structures in SAbDab contain only these fragments.
- a numbering scheme? - a system to annotate equivalent positions in antibodies. A scheme can often be applied by examining the sequence of the antibody only.
FAQ
How do I:
- Go to the summary page for the structure as described above. (example)
- Click on the "Downloads" tab. Four files will be available:
- the structure in PDB format as deposited. (example)
- the structure in PDB format with the antibody chains numbered using Chothia numbering. (example)
- the structure in PDB format with the antibody chains numbered using IMGT numbering. (example)
- a csv (values separated by tabs) summary file containing annotations pertaining to the structure. (example)
- Simply click on each link to download the file. See the "Data and Downloads" section below for more information about file formats.
- Go to the Search Structures page, click on the "Get all structures" tab and press the button. A table with all the current entries will be displayed.
- Go to the Search Structures page and click on the "Search structures by attribute" tab.
- Select from a number of properties including:
- Experimental method (e.g. X-Ray, NMR etc)
- Bound state and, if bound, antigen type (e.g. protein, hapten etc)
- Antibody species
- A list of structures that satisfy the conditions will be returned.
- Here you go!
- We hope this to serve as an antibody-antigen docking benchmark resource.
- Narrow your search (e.g constrain by antigen type) by searching by attribute on the Search Structures page.
- Here you go!
- Single-domain antibodies can be selected by choosing the antibody type 'VHH' on the database search forms.
- Go to the Search Structures page and click on the "Search for a non-redundant set of antibodies" tab.
- Select the properties that you wish the structures to have, e.g. whether only bound antibodies should be considered and structure quality cutoffs.
- Select the sequence-identity threshold for at which antibody sequences should be clustered at (variable domains only).
- Note: this may a particularly useful tool for studying antibody interactions and docking protocols. (e.g. 6% of protein antigens in SAbDab are lysozyme!)
- A non-redundant list of structures will be returned upon submission.
- Select a dataset as described above. You will be presented with a list of structures.
- The data for each may be downloaded individually using the links in the rightmost column of the table.
- The whole dataset may be downloaded as a zip file by clicking on the link at the bottom of the page. This will create an archive file which must be downloaded within 20 minutes of creation.
- Alternatively, use our download script to download the data at your leisure (Help?).
- Go to the Search by Sequence Similarity page.
- Paste the sequence of the antibody you wish to find structural templates for in the text boxes. This can be either a heavy chain or a light chain or both.
- Choose the number of structures that should be returned (between 1 and 100).
- Click "Search database" to return a list of structures with the highest sequence identity to your antibody (variable region only).
- Scroll down the page to view an annotated alignment between your sequence and each template.
- By default sequence identity is calculated over the variable region of the antibody. Users may also choose different regions to calculate the identity over (e.g framework or CDRs).
- Go to the Search CDRs page and click on the "Search CDRs by attribute" tab.
- Choose which CDR definition to use (Chothia, Kabat, Contact, IMGT, or North)
- Choose the type of CDR you wish to select (H1, H2, H3, L1, L2 or L3). Leave as "All" to select all.
- Choose the length the CDRs that should be returned. Leave blank to select all.
- Choose the other attributes that the structures should have.
- Click "Get CDRs" to return a list of CDR structures.
Data and Downloads
Available Data
For each entry in SAbDab the following data is available for download:
- The structure file in PDB format as deposited to the protein data bank. (example).
- Renumbered structure (Chothia and IMGT) files in PDB format. (example).
- All chain identifiers are retained.
- Chain pairings (heavy-light-antigen) are recorded in a REMARK record in the header. e.g. for structure 1ahw:
REMARK 5 PAIRED_HL HCHAIN=B LCHAIN=A AGCHAIN=C AGTYPE=PROTEIN
REMARK 5 PAIRED_HL HCHAIN=E LCHAIN=D AGCHAIN=F AGTYPE=PROTEIN
- Variable region of the chains are Chothia (or IMGT) numbered. e.g CA atom at Chothia position H82A on chain B in structure 1ahw:
ATOM 3877 CA SER B 82A -18.113 15.679 27.979 1.00 6.53 C - Residues outside this region are numbered sequentially.
- "Non-antibody" chains retain their original numbering.
- A summary file in csv format (values separated by tabs). (example).
- The first row of the file is a header containing the name of each field.
- Each subsequent row corresponds to a heavy-light chain pairing and associated annotations. e.g. for structure 1ahw the first 6 fields are:
pdb Hchain Lchain model antigen_chain antigen_type ...
1ahw B A 0 C protein ...
1ahw E D 0 F protein ...
- To view in excel or open office, open the file and when prompted choose "separated by Tab".
Download methods
When any dataset (list of entries) is selected in SAbDab there is an option to download the data using two methods:
- Download an automatically generated zip file (preferred).
- In the sidebar, click "Downloads".
- Click on the link to download the zip file. A zip file will be created and automatically downloaded that contains the files described above for each selected structure.
- This file will be available for 20 minutes after creation.
- Download using the SabDab download script.
- Download the summary file for your selection.
- From a unix command-line run the script, specifying the data that you wish to retrieve. e.g. Download the original structure and the imgt annotations for the structures in the summary file:
$ python sabdab_downloader -s summary_file.csv -o path/to/output/ --original_pdb --imgt - This will create a folder called "sabdab_dataset" in the folder "path/to/output/". Within that will be a directory for each entry in the summary file containing the requested data.
- Functionality has only been tested in linux. Please use the zip file approach for all other operating systems.
Summary file fields
A summary file is created when a dataset is selected in SAbDab and is available for each structure individually. Each row corresponds to a heavy-light chain pairing in a PDB structure. Each pairing is annotated with the following fields.
| PDB | The PDB accession code (e.g. 12e8) |
| Hchain | The chain identifier for the heavy chain (e.g. "H"). This is "NA" if the light chain is unpaired. |
| Lchain | The chain identifier for the light chain (e.g. "L"). This is "NA" if the heavy chain is unpaired. |
| model | The model identifier for the pairing (e.g "0","1","2"...). This is "0" for X-Ray structures ) |
| antigen_chain | The chain identifier for the bound antigen chain (e.g. "A"). If the antigen has multiple bound antigen chains, these are separated by a "|" (e.g "X | Y"). For non-polymer antigens this refers to the chain identifier of the corresponding HETATM records (i.e. it may be the same as either the heavy or light chain identifier) |
| antigen_type | The classification of the antigen. Either: protein, peptide, carbohydrate, nucleic acid or hapten. This is "NA" if the heavy-light pairing is unbound. |
| antigen_het_name | The HETATM of the antigen if it is non-polymer. This is "NA" if the antigen is a polymer or the heavy-light pairing is unbound. |
| antigen_name | The name of the antigen. This is "NA" if heavy-light pairing is unbound or "?" if unknown. |
| short_header | The short header of the structure. Typically a short description of the type of molecule in the structure entry. |
| date | The deposition date of the structure to the PDB. |
| compound | The description of the molecule in structure. Typically the title of the associated publication. |
| organism | The organsim(s) of the molecule(s) in the structure. |
| heavy_species | The species of the heavy antibody chain. If it is from multiple species (e.g. Chimeric or Humanized) these will be separated by a single comma. |
| light_species | The species of the light antibody chain. If it is from multiple species (e.g. Chimeric or Humanized) these will be separated by a single comma. |
| antigen_species | The species of the antigen chain. If it is from multiple species these will be separated by a single comma. |
| authors | The authors of the structure. |
| resolution | The resolution of the structure if determined by X-Ray diffraction or Electron Microscopy. |
| method | The method with which the structure was determined. |
| r_free | The Rfree value of the structure if determined by X-Ray diffraction. |
| r_factor | The R factor value of the structure if determined by X-Ray diffraction. |
| scfv | Whether the structure is a single chain Fv. True or False. If true, the heavy and light chain identifiers may be the same depending on how the structure has been deposited. |
| engineered | Whether the structure has been engineered. True or False. |
| heavy_subclass | The IMGT variable subgroup of the heavy chain. Structures that are not available in the IMGT database have a subgroup assigned by SAbDab. |
| light_subclass | The IMGT variable subgroup of the light chain. Structures that are not available in the IMGT database have a subgroup assigned by SAbDab. |
| light_ctype | The type (Kappa or Lambda) of the light chain. |
| affinity | The affinity of the antibody to the antigen present in the structure (KD - M). |
| delta_g | The ΔG of the antibody to the antigen present in the structure (kcal/mol). This has been manually calculated. |
| affinity_method | The method by which the affinity data was collected (SPR, ITC or other). |
| temperature | The temperature at which the affinity data was collected (°C). |
| pmid | The pubmed identifier that is the source of the associated affintity data. |
Contact
Feel free to contact us at opig <~at~> stats.ox.ac.uk for any issues, misannotations or general enquiries about SAbDab.
