About Thera-SAbDab
The Therapeutic Structural Antibody Database (Thera-SAbDab) is a database of immunotherapeutic variable domain sequences and their corresponding structural representatives in SAbDab (which harvests data from the PDB). It updates structural mappings alongside SAbDab on a weekly basis. It detects not only exact sequence matches to known structures, but also close sequence matches (divided into two categories: 95-98% seqID, or 99% seqID).
We update Thera-SAbDab whenever a new WHO International Non-proprietary Name (INN) list is released, adding all therapeutics with an accompanying variable domain sequence. We also update the clinical trial status of all actively-developed therapeutics according to the latest updates on AdisInsight. We host this up-to-date list of therapeutic sequences with metadata on the Thera-SAbDab search page.
Thera-SAbDab has been built by the Oxford Protein Informatics Group (OPIG) under an open-innovation agreement.
If you use this tool, please cite our paper: Raybould, M.I.J., Marks, C. et al. (2019) Nucleic Acids Res. gkz827
If you believe we have missed a WHO-recognised therapeutic with a public sequence, please contact us at opig <~at~> stats.ox.ac.uk
Example Queries
Scenario 1: I'm developing a biosimilar for certolizumab, and want to see if there are any publicly-available structures of the parent antibody.
-> Search Thera-SAbDab by INN: "certolizumab"...
Returns the summary...
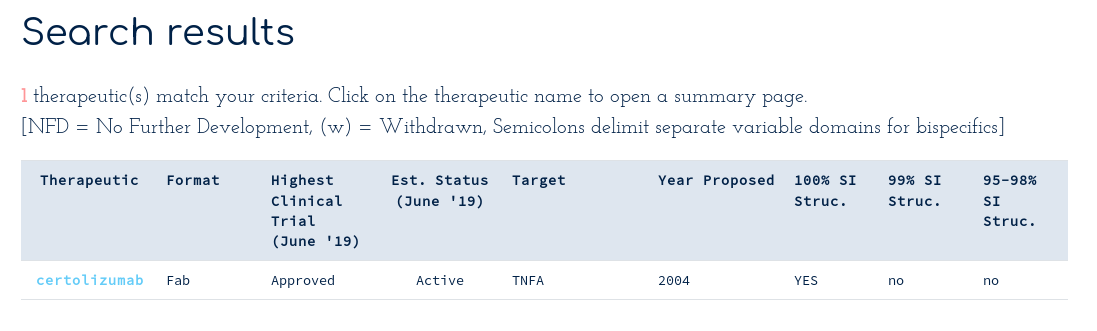
This shows that there is at least one sequence identical structure of certolizumab in the PDB. Clicking on the Certolizumab link takes you to the summary page...
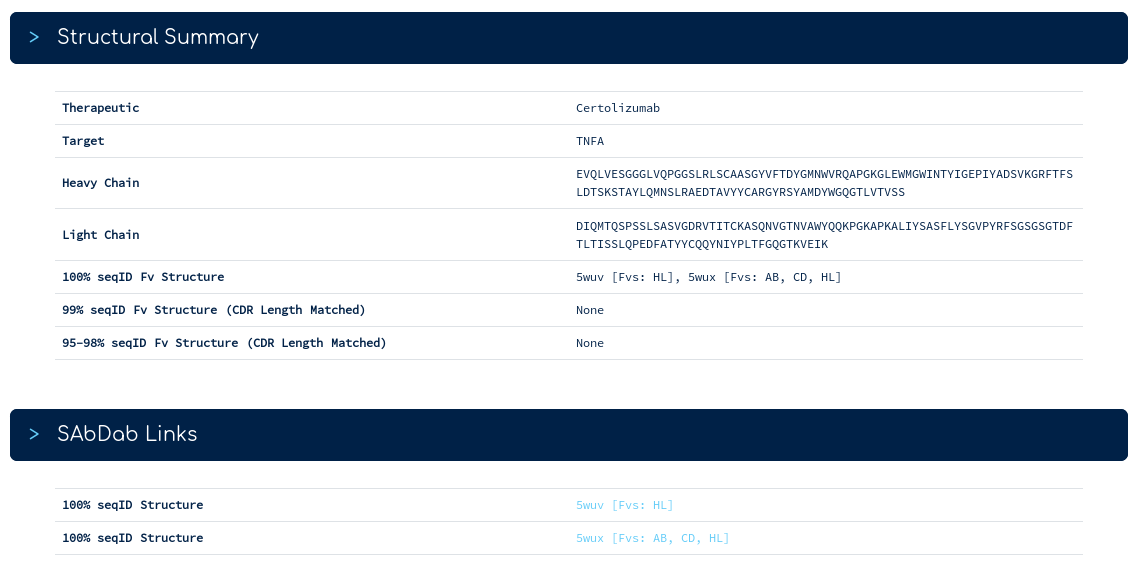
The top of the summary page gives you our database sequence for certolizumab alongside its structures. There are two sequence identical PDB entries. One with a single Fv region, one with three Fv regions. The links below take you into the SAbDab entry for each PDB. Following the link for 5wux...

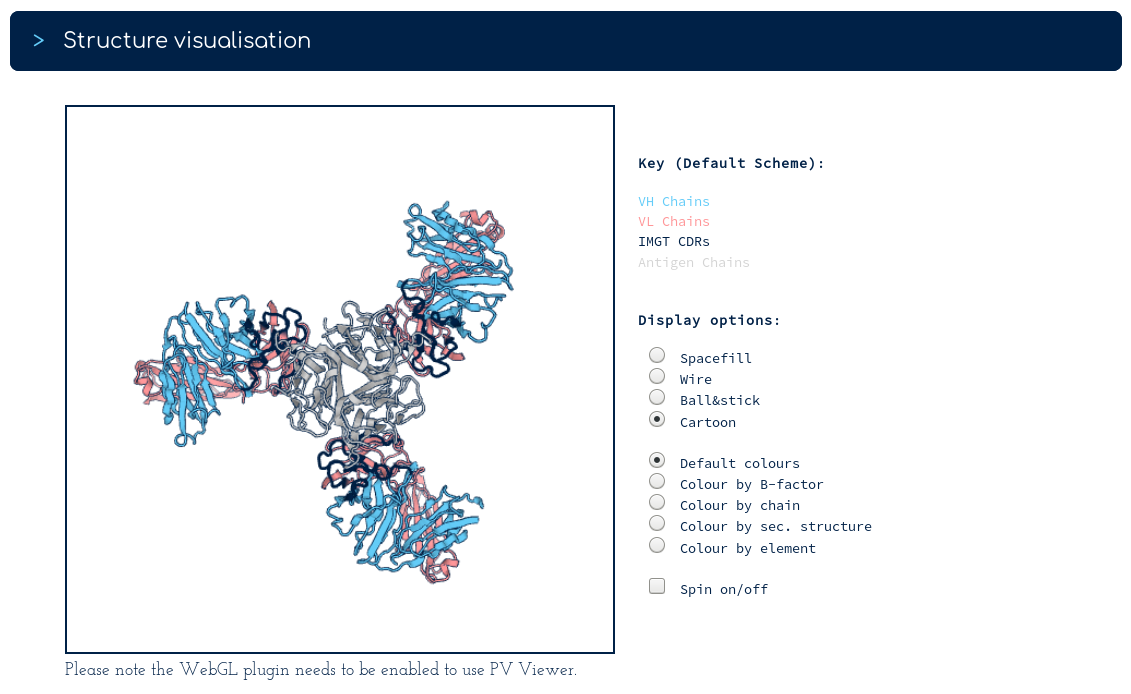
We can see that 5wux is a trimeric bound structure. Also on the certolizumab summary page...
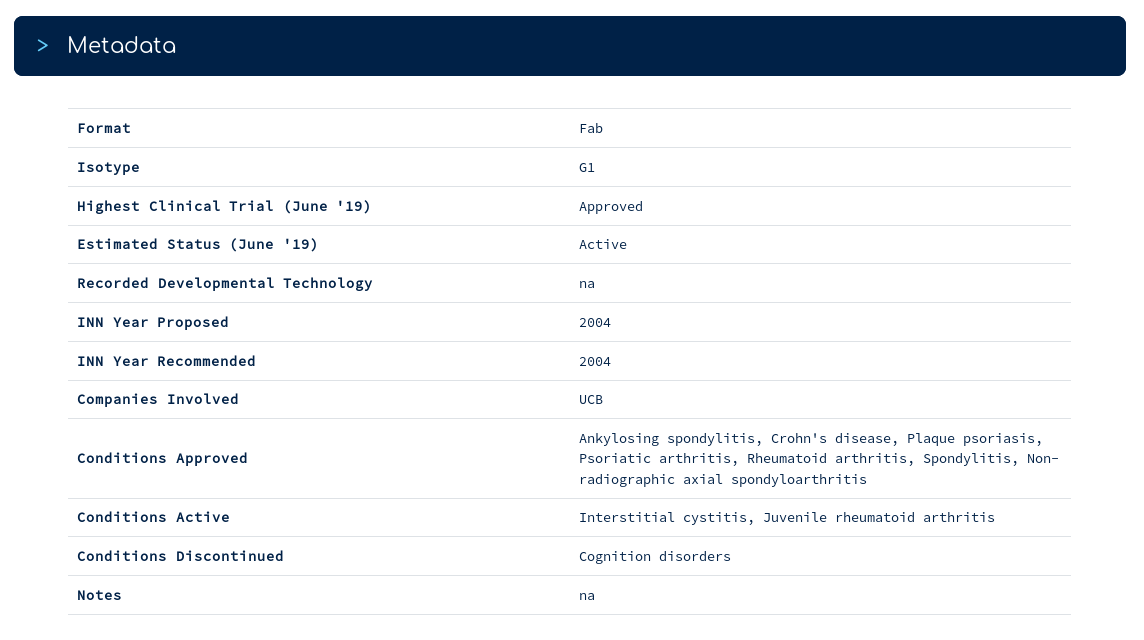
...is a series of metadata for certolizumab. For example, you can see which conditions it is currently approved/active for, and which it has been discontinued against (according to AdisInsight).
Scenario 2: Having found that there is a structure for Certolizumab (a TNFalpha inhibitor), I'm interested in the general structural characteristics of TNFalpha binding drugs.
-> Search Thera-SAbDab by attribute: Target = "TNFA"...
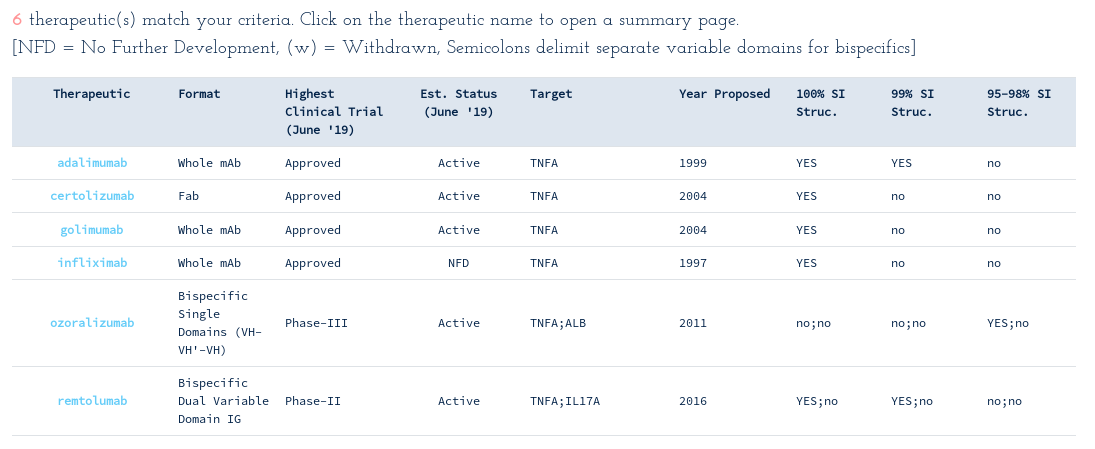
This reveals there are six WHO-recognised antibody- and single-domain antibody-derived therapeutics designed to bind to TNFA, four of which are monoclonal and two bispecifics. Most therapeutics have publicly-available crystal structures for their TNF binding variable domains. Follow the SAbDab links on each summary page to inspect and download your pre-numbered structures of interest.
The fact that one therapeutic targets IL17A and TNFA simultaneously catches your attention and you follow up to see if we know any structures of IL17A binders by using another attribute search...
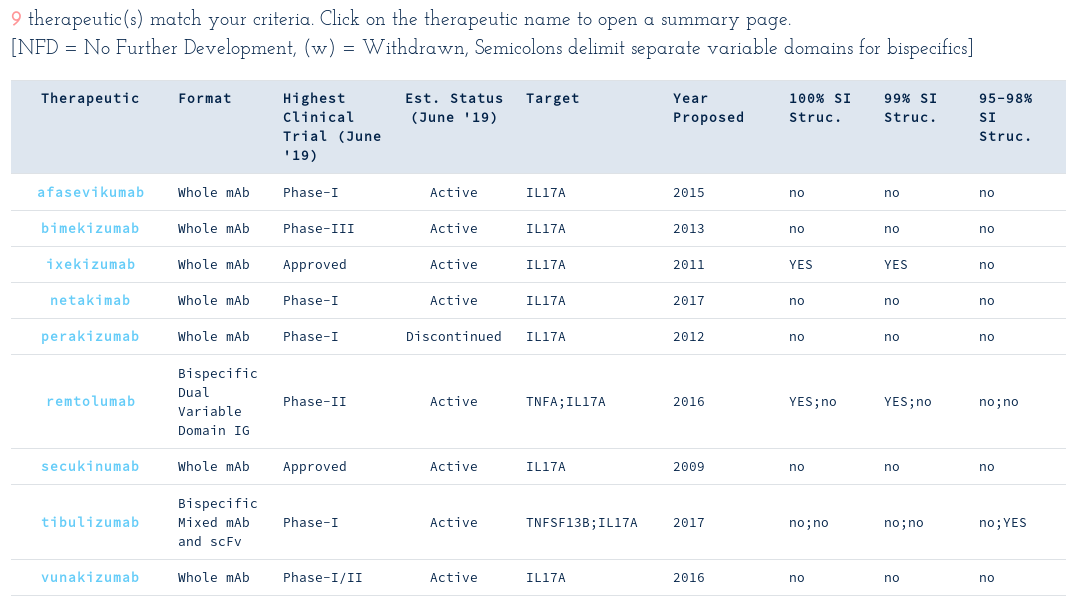
As of early June, none of the nine WHO-recognised IL17A binders had any publicly-available crystal structures. However, on 19th June 2019, Eli Lilly released a structure for ixekizumab (6nov), and a near structure for tibulizumab (6nou)!
Scenario 3: You believe you have found a completely novel anti-TFNA antibody and want to check to see how similar it is to other known therapeutics.
-> Search Thera-SAbDab by Fv sequence identity:
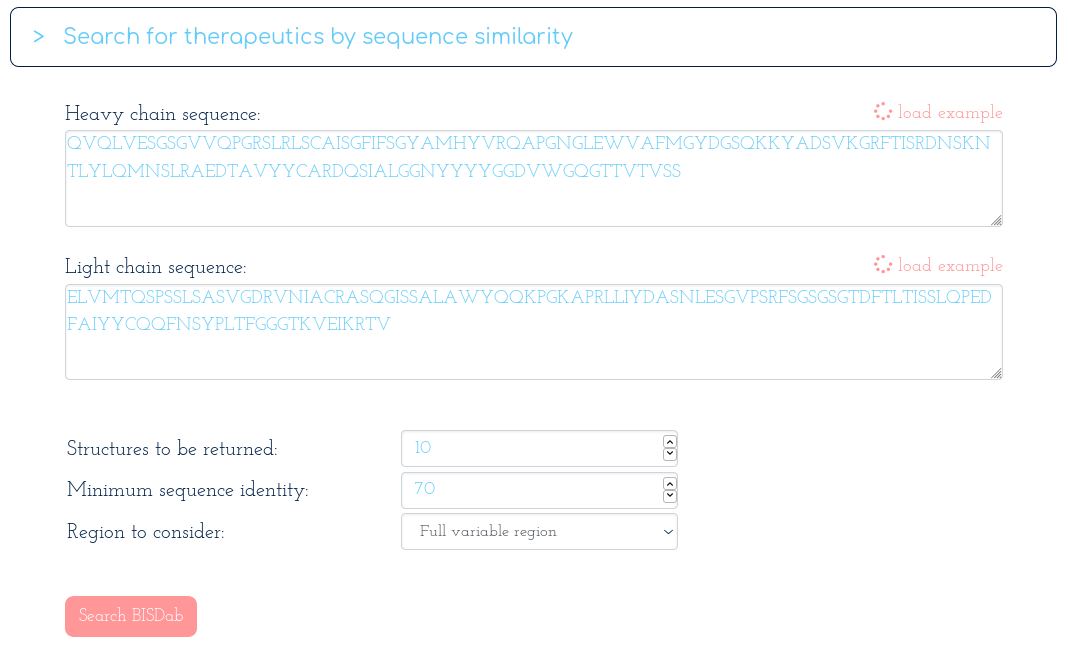
With the following table of results...
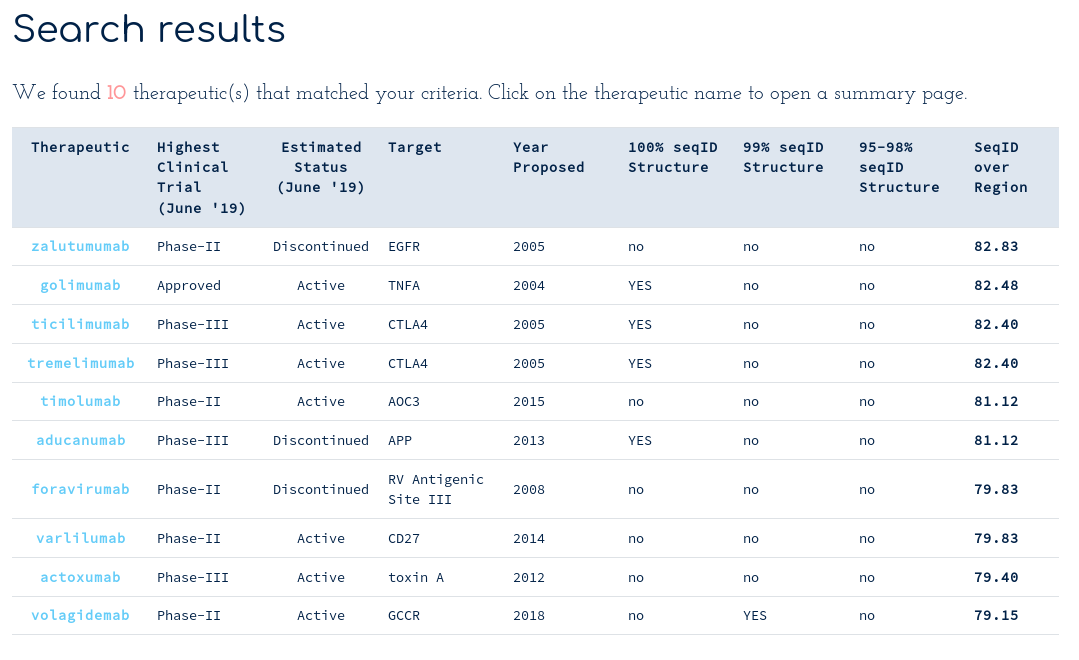
There are at least ten therapeutics within 70% sequence identity of your query across the whole Fv region. You notice Golimumab, another TNFA-binding antibody, has 82% sequence identity to your submission. You scroll down to the alignments section...
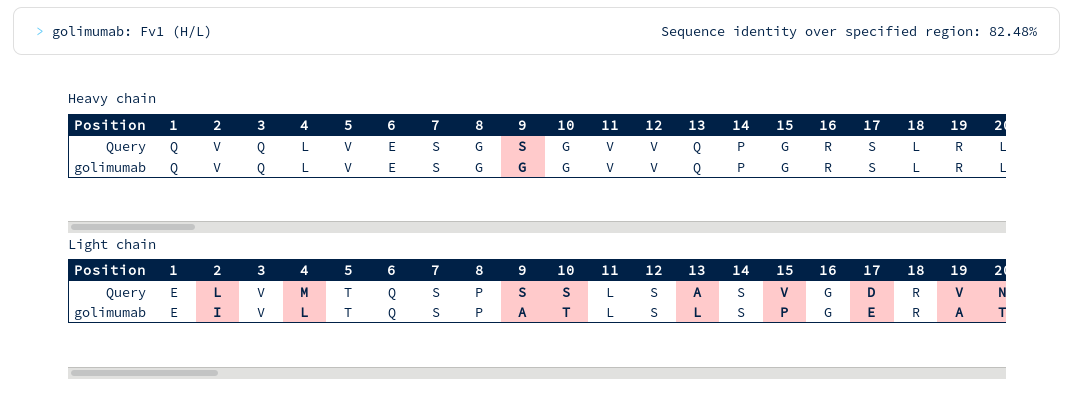
There appear to be many more mutations in the light chain than in the heavy chain. You're interested in precisely how similar the heavy chain binding site might be to Golimumab, so you run another search for similarity across just the heavy CDR residues...
Golimumab is again within your required threshold, with the following alignments to CDR regions...
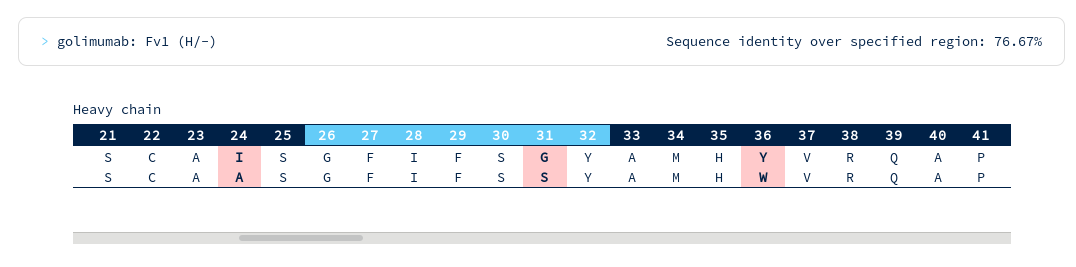
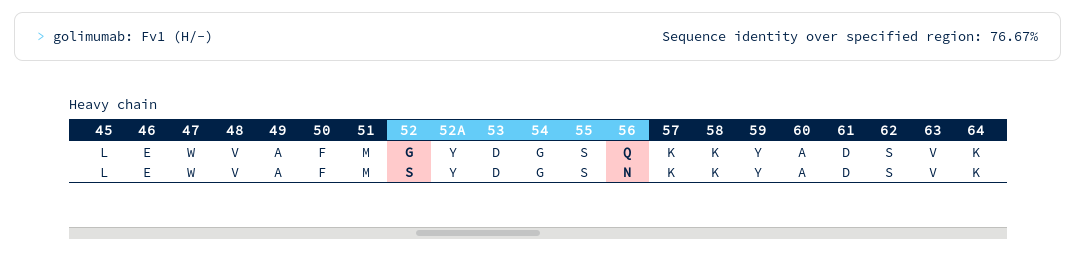
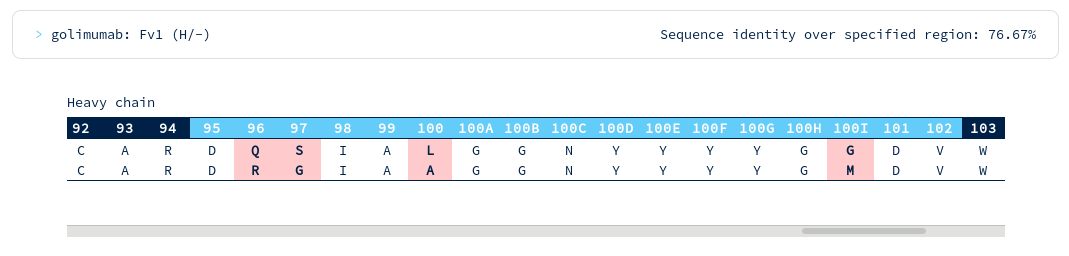
There are surprisingly few heavy chain CDR mutations away from Golimumab (particularly in the CDRH3 region). You follow the link to the Golimumab summary page, and from there to the 100% sequence identical structure (5yoy). Looking at its binding mode...
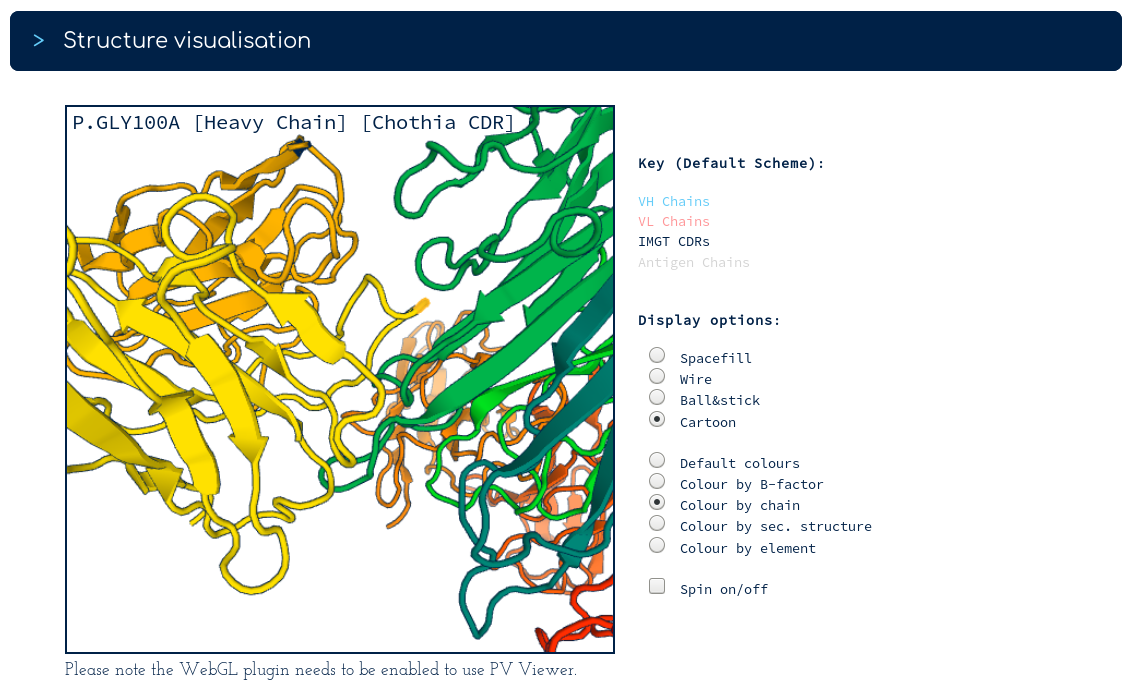
...binding to TNFA (green) seems to be dominated by the heavy chain (yellow). This would result in less selection pressure on the light chain (orange), beyond being compatible with the TNFA-binding heavy chain. You conclude that the binding mode of your antibody probably shares many characteristics with a currently-approved TNFA therapeutic.
Contact
Feel free to contact us at opig <~at~> stats.ox.ac.uk for any issues, misannotations or general enquiries about Thera-SAbDab.
