Search PLAbDab by keyword to find HIV-related antibodies using a regular expression: "hiv|immunodeficiency" ...

Returns the following results...

This shows a list of entries from different data sources that contain your keywords in their title. Only one entry will be shown per data source, but information from all entries can be downloaded using the "Download" link
Clicking on the "Source URL" link will lead you to where the sequences were scraped from. More details on the antibodies target can often be found there. Clicking on the ”More info” link will lead you to a page with more information on the selected entry.
The top of the "More info" page contains metadata for the selected entry, like the VH and VL sequences, and the organism. For example, here we can see that this is a human antibody.
The second section of the page shows the structure of a three-dimensional model predicted using ABodyBuilder2:
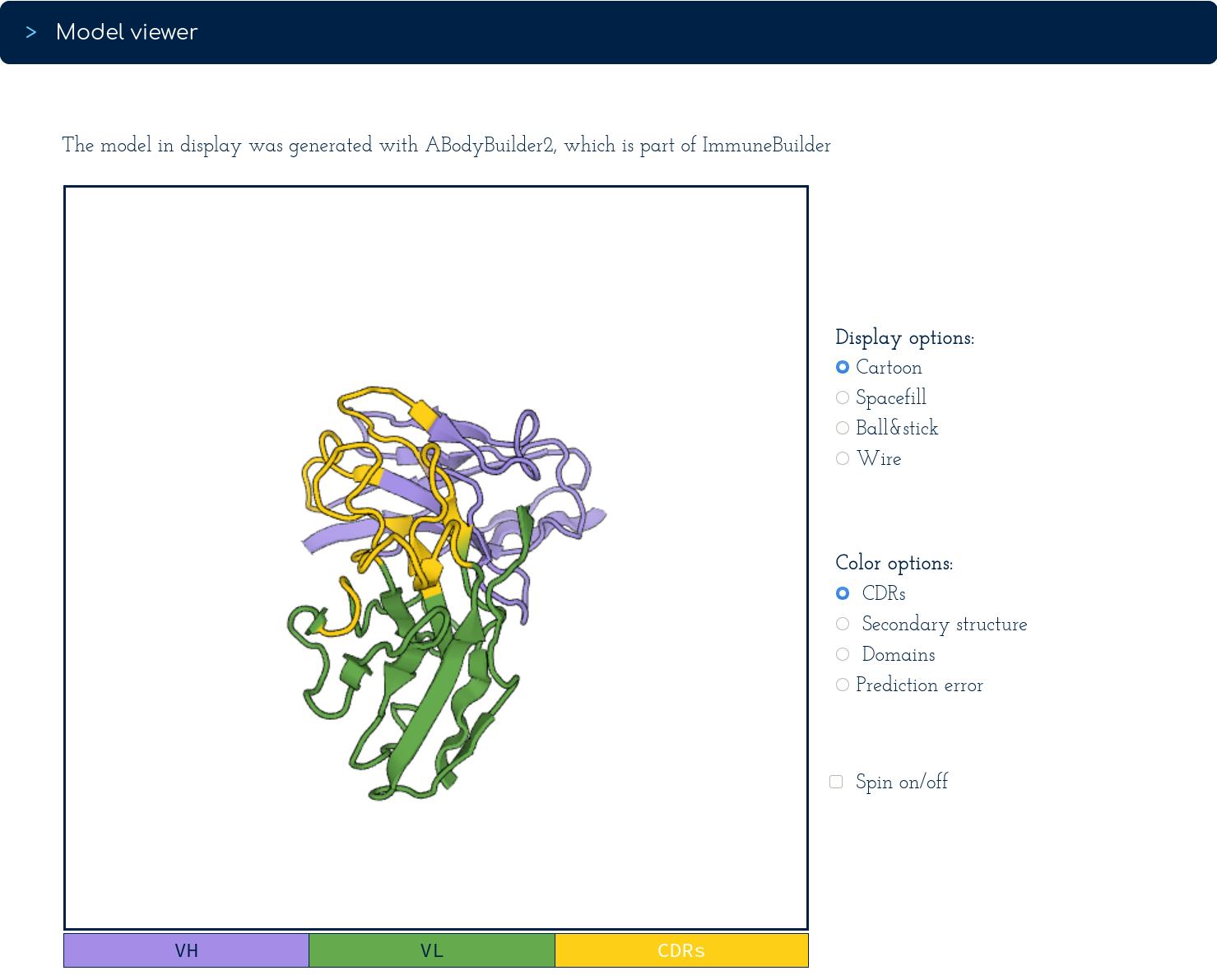
In the viewer, the structure can be represented using a selection of settings, such as, cartoon (shown above), space-fill, ball&stick or wire. To facilitate manual inspection of the antibody model, its structure can also be coloured by secondary structure, domains, or predicted error.
The bottom part of the page presents the user with options to download the structural model, or details on the retrieved antibody:
To find information on the query antibody, we can search PLAbDab using sequence identity or structural similarity. We will first search for the 20 entries with highest average sequence identity over the VH and VL, above the specified cutoff (80%).
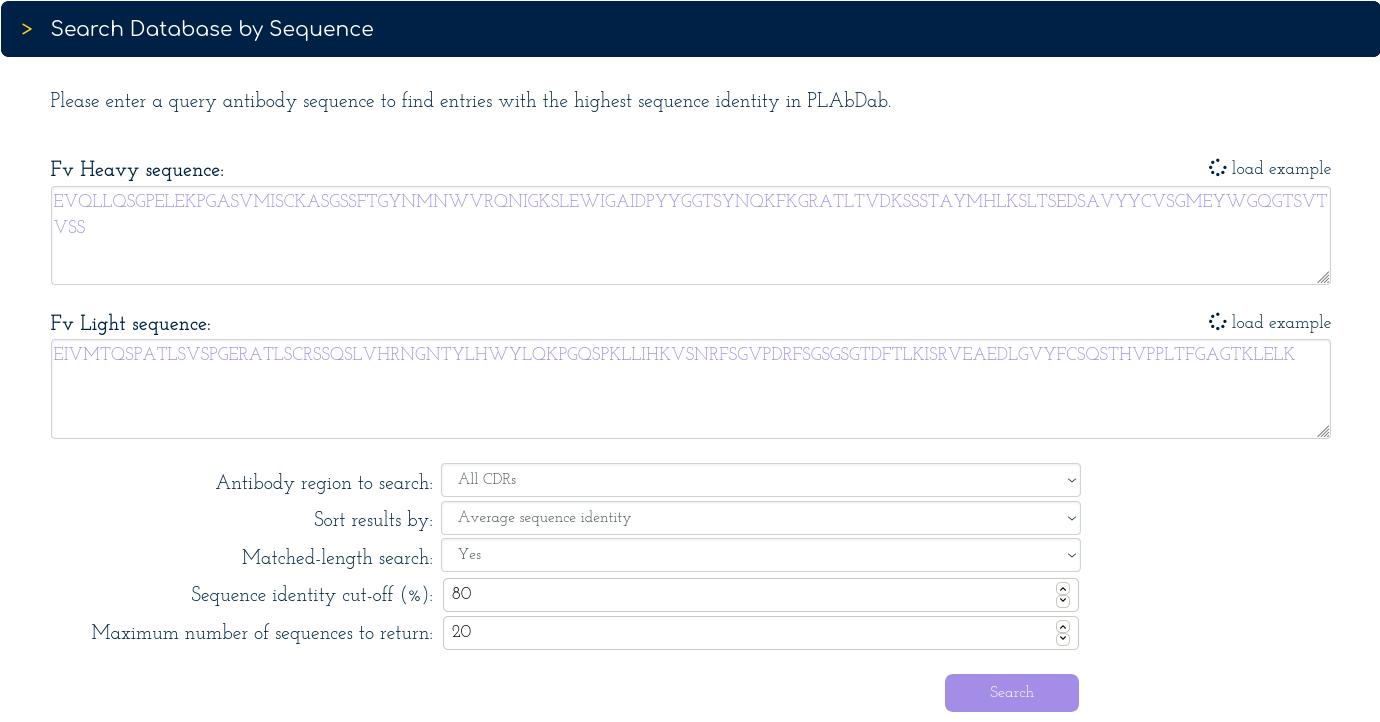
This reveals 20 antibodies with an identical sequence over the 6 CDR loops from several sources:
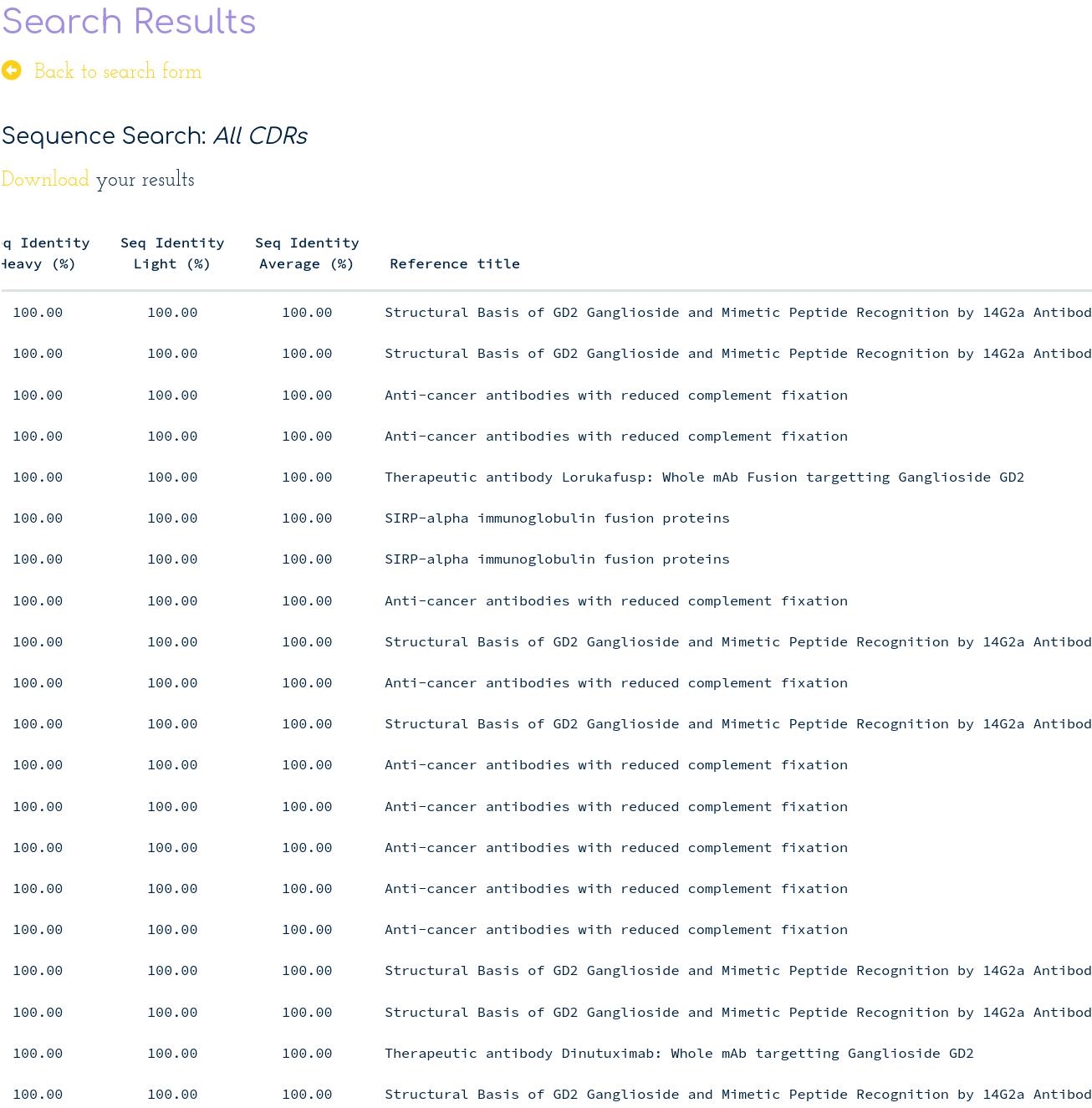
The source with most entries is a patent describing ”Anti-cancer” antibodies. This already gives us some information on what the function of this antibody may be. The second source with most entries corresponds to a study that describes a GD2 Ganglioside antibody, and that deposited 5 crystal structures in the PDB (4tuo, 4tuj, 4tul, 4trp, 4tuk). Another retrieved entry of interest is the therapeutic antibody “lorukafusp”, which targets the same antigen as “dinutuximab”.
PLAbDab can also be searched for antibodies with a similar structure over the CDR loops. Structure search will occasionally reveal functional links that would not be found by using sequence search alone. We will search PLAbDab for the 20 antibodies with lowest CDR RMSD to a model of the query, below the specified cutoff (3Å).
This reveals the following 20 entries in PLAbDab:
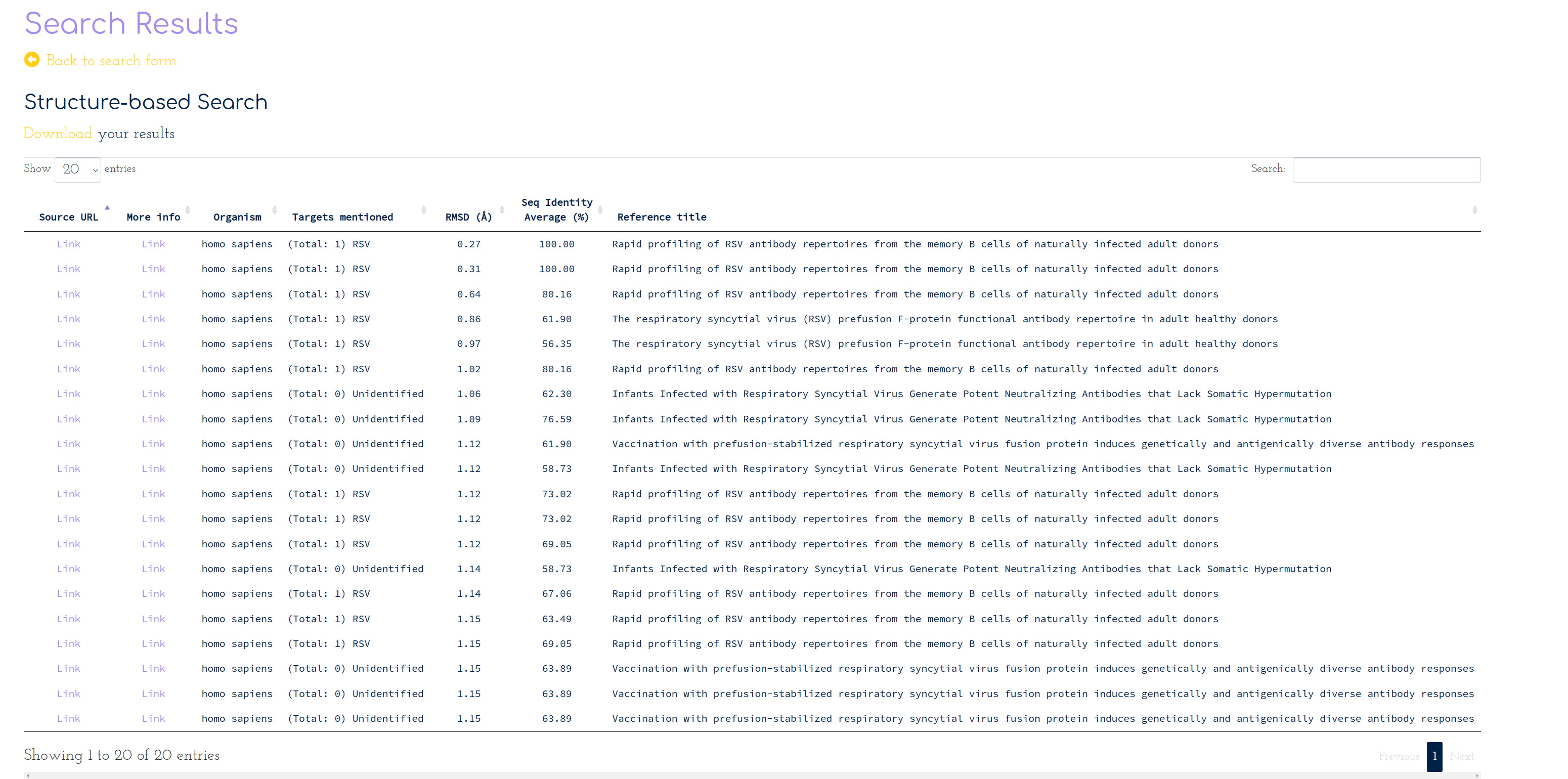
In this case, the results returned by the structure and sequence searches agree well. Although in this case structure search does not provide much additional information, it can be used to validate results from sequence search.
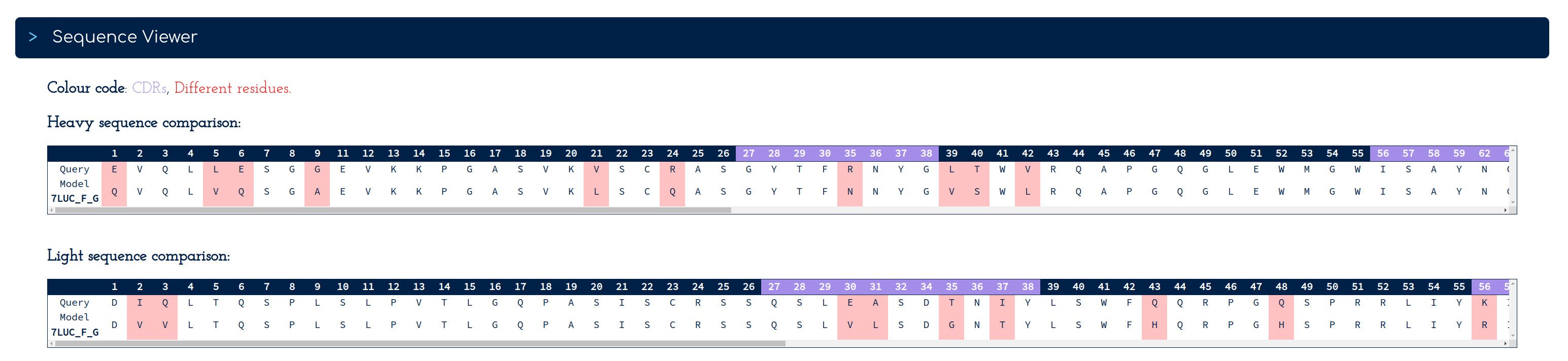
As an extra feature, the More Info pages for sequence and structure search results present two sequence viewers to facilitate a quick comparison of the VH and VL sequences corresponding to the user query and the sequences in the reference outputted by PLAbDAb, highlighting the position of CDRs and residue differences.
Feel free to contact us at opig <~at~> stats.ox.ac.uk for any issues or general enquiries about PLAbDab.
Brennan Abanades et al. “The Patent and Literature Antibody Database (PLAbDab):
an evolving reference set of functionally diverse, literature-annotated antibody sequences and structures”. In: Nucleic Acids Research, Volume 52, Issue D1, 5 January 2024, Pages D545-D551 [ link]
Download BibTex Reference
Header image credit: David Goodsell