Current Research
We carry out research on many topics in protein informatics, including the following areas. Please use the thumbnails to navigate to summaries of the research by group members in each area.
Protein Structure

Alexi (Hussain) Siddiqui (DPhil)
Understanding protein function requires the probing of both structure and dynamics. Traditional methods have limitations when attempting to capture dynamic behaviour. Hydrogen-Deuterium Exchange Mass-Spectrometry (HDX-MS) quantitatively assesses conformational dynamics and empirical models have been used to link the data to molecular dynamics (MD) simulations, potentially offering a more complete view of protein behaviour. My research is focused on reliable and robust methods for generating accurate conformations relevant with respect to experimental HDX-MS data. This is relevant to recently released structural prediction models. Using the wealth of existing data, we want to apply these methods for new insights. All developed code will be released, readily adaptable to existing analysis pipelines.
.jpg)
Qurat ul ain (Annie) (DPhil)
Current approaches for protein design require multiple iterations of the design-make-test experimental cycle and provide limited control over the properties of the resulting molecules. I’m developing deep learning-based methods for designing de novo proteins with specific physicochemical properties. Computationally, this becomes a multi-objective optimisation problem where the output must be novel, diverse and physically plausible - how exciting! Feel free to reach out if you’re interested in similar topics and would like to have a chat.

Gabriel Abrahams (DPhil)
Proteins are remarkable nano-machines that carry out the myriad functions required for life to exist. Engineering proteins to have novel functions has a vast range of applications, ranging from medical developments such as combating anti-microbial resistance, to climate friendly industrial manufacturing. In my DPhil, I am working to develop a machine learning pipeline to steer directed evolution: a method for utilising the power of natural evolution to produce proteins with desirable capabilities that were not required to survive in nature. These experiments will be performed in the lab, in a massively high throughput screening platform currently being developed by the Engineered Biotechnology Research Group at Oxford.

Aaron Maiwald (DPhil)
My research focuses on understanding how neural networks process and learn from genomic data. Genomic language models such as the Nucleotide Transformer have been pretrained on vast amounts of DNA data and show promising performance on a range of benchmarks. As these systems become increasingly powerful and widely used in biological research, it's crucial to understand exactly how they arrive at their predictions. Using techniques from mechanistic interpretability - an emerging field that reverse-engineers neural networks - I develop methods to reveal how these systems represent and transform biological information. This work is aimed to help us build more reliable and capable AI systems in genomics, and biology more broadly. If any of this interests you, please feel invited to reach out!
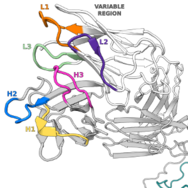
Immunoinformatics

Matthew Raybould (Postdoc)
My research applies immunoinformatics to improve therapeutic design and to better our understanding of the immune response. During my DPhil, I captured and compared structural representations of therapeutic and natural antibodies, leading to new structure-aware approaches for in silico developability assessment and screening library design. My research is now focused on incorporating structural awareness to improve our ability to identify broader sets antibodies with functional commonality and to define the functional boundaries of different classes of adaptive immune receptor.

Nele Quast (DPhil)
I develop and train deep learning models for T-cell receptor structures. I'm interested in training models that retain equivariance, merge sequence and structure information and can be injected with conditions or constraints. I'm also interested in the structure of the interface between TCRs and their pMHC antigen. Beyond my research I'm passionate about improving gender representation in STEM and have acted as president of the Oxford Wom*n in CS society.

Oliver Turnbull (DPhil)
For an antibody to make an effective therapeutic, it must both bind to its target and be free from developability issues, such as aggregation, poly-specificity, and poor expression levels. By either limiting our search space to developable antibodies, or building methods to engineer out developability issues, the success rate of therapeutic antibodies can be increased. My DPhil aims to tackle both problems using generative machine learning methods.

Fabian Spoendlin (DPhil)
Antibodies are an important component of the immune system and are increasingly used as therapeutics. Recent advances in protein structure modelling make it possible to accurately predict the structure of antibodies from their amino acid sequence. A limitation of current structure prediction tools is that they only predict the structure of a single conformation of an antibody. However, antibodies are flexible molecules that frequently transition between a set of distinct structural conformations and flexibility is key to many functional properties. During my DPhil, I aim to develop antibody structure prediction tools that capture the flexibility of antibodies and predict the structure of multiple conformations.

Benjamin McMaster (DPhil)
T cells are a key part of our immune system, responsible for fighting pathogens and regulating immune responses. To identify foreign invaders T cells, use their receptors (TCRs) to rapidly screen and identify antigens. Although, key to our health and survival, the map between TCR composition and antigens is still poorly understood. I aim to apply newly develop deep learning models in protein structure prediction to TCR data to better understand the rules that govern antigen-specific T cell response.

Henriette Capel (DPhil)
Antibodies are an important class of biotherapeutics. The process of engineering a therapeutic antibody is time and cost intensive, with many of them failing due to developability issues, such as low expression, low solubility, and high aggregation. My research aims to develop computational methods to improve the outputs of antibody developability workflows. These tools will incorporate experimental data including negative data, which is essential to guide the antibody engineering process.

Isaac Ellmen (DPhil)
Antibodies work by binding to their targets (antigens), and either inhibiting their function or activating other components of the immune system. Predicting the mode by which an antibody binds to its cognate antigen is called antibody-antigen complex modelling or docking. While general protein complex prediction has seen great improvements in recent years, driven by methods such as AlphaFold Multimer, antibody-antigen complexes are still difficult to model because we rarely have useful homologs to provide co-evolutionary information. My DPhil project is focused on developing new machine learning docking models to more accurately predict antibody-antigen complexes.

Odysseas Vavourakis (DPhil)
I develop geometric generative models for protein structure, sequence, and dynamics, with a focus on de novo antibody design. The challenge is to generate human-like antibodies that specifically bind to a target while meeting a range of ancillary constraints—many related to antibody flexibility—in order to create viable therapeutics. I am therefore particularly focused on developing machine learning models to accurately predict conformational ensembles and enable robust antigen-conditional design. An interesting recurring challenge in the antibody space is the comparative scarcity of data compared to general-protein systems.

Marius Urbonas (DPhil)
Large deep-learning foundation models have become the dominant paradigm across many fields. Their ability to extract useful representations allows for efficient finetuning to target tasks. Even more interestingly we can predict exactly how the performance of these models will improve as we increase the model size or the amount of data without even training them, which motivated the building of ever larger language and vision models. I am interested in whether similar scaling laws can be found for immunology task modelling. My work aims to establish the data requirements for reliable immune modelling and guide more efficient therapeutic antibody discovery.

King Ifashe (DPhil)
T-cell receptor (TCR) cross-reactivity poses a challenge in developing safe and effective immunotherapies, as TCRs can inadvertently recognise multiple peptides presented by the Peptide-human leukocyte antigen (pHLA) proteins, leading to off-target effects. I work on studying how the structural diversity of these peptides influences TCR binding and cross-reactivity by developing and applying computational tools that identify common conformational patterns. This work aims to inform the engineering of safer, more specific TCR-based therapies that minimise off-target effects.
Small Molecules

Garrett Morris (Associate Professor)
My primary focus is on methods development in computer-aided drug discovery, chiefly in high throughput docking, ligand-based virtual screening, network pharmacology, cheminfomatics, bioinformatics, machine learning and more recently protein engineering. Current research projects include: addressing the limitations of scoring functions in docking, in particular to improve our understanding of molecular recognition of small molecules; handling receptor flexibility in protein-ligand docking; and fragment-based drug discovery.

Fergus Imrie (Florence Nightingale Fellow)
My research develops and applies novel machine learning methods to challenges in healthcare, medicine, and drug discovery. Methodologically, my work spans both predictive and generative methods, explainable AI, feature selection, causal inference, and learning from unlabelled data. From an application perspective, I am currently particularly interested in structure-based drug design.

Martin Buttenschoen (DPhil)
I am interested in machine learning models that predict protein-ligand binding. Currently I am working with graph neural networks and generative models.

Kate Fieseler (DPhil)
A fragment screen offers an information rich starting point for derivative compounds that can recapitulate fragment interactions. I am interested in how to maximally explore the fragment merge-design space with in silico design. By prioritizing synthetic tractability, fragment derivate designs can be synthesized via high-throughput multi-step chemistry reducing the overall cost and time to experimentally test them. In collaboration with XChem at Diamond Light source, I work directly with organic chemists and structural biologists to drive their fragment development projects forward.

Isak Valsson (DPhil)
My research interests involve developing robust machine learning methods for early drug-discovery problems. Recently, I’ve been working on developing structure based scoring functions that work better in an out-of-distribution (OOD) setting, i.e. when the training data occupies a different area in chemical space than the test data. Additionally, I’ve been exploring different ways to benchmark the OOD performance of binding affinity predictors, and different ways of featurising bound protein-ligand complexes.

Alexander Hasson (DPhil)
The Eric O'Neill lab recently identified a combination of biologically-targeted agents, that could restore epigenetic control and revert squamous pancreatic cancer cells to a more regulated differentiated state. Although we observe a shift towards normal cell epigenetics, this drug combination is not fully optimised to drive the phenotypic shift. In my research, I am developing and aim to use an innovative data-driven artificial intelligence (AI) approach to delineate novel molecules that can demonstrate improved and robust re-normalisation of epigenetic status and pancreatic differentiation, in order to offer new treatments for this hitherto intractable cancer. A major part of this work is the development of an inverse screening protocol, in order to find the protein targets of small molecules.

Yael Ziv (DPhil)
My current research interests lie in structure-based drug design, where the primary focus is on developing small molecules that have a strong and specific affinity for a particular 3D protein structure. My research focuses on deep-learning generative models, exploring innovative approaches to integrate targeted protein information into the design process. I am particularly interested in the necessity for novel deep-learning methods that can effectively incorporate the targeted protein, all while prioritizing the generation of molecules that are both chemically and physically plausible.

Adelaide Punt (DPhil)
My research examines the impact of noise on small molecule activity prediction, identifying robust pairings of model and molecular embedding under increased artificial noise. By clustering molecules into chemical domains, I introduce domain-specific noise to mimic real-world variability. Within a federated learning framework, I address challenges from heterogeneous data distributions and client-specific noise variability by applying noise mitigation methods across both pre-processing and training steps, aiming to remove and smooth experimental noise.

Arun Raja (DPhil)
My DPhil research involves the development of geometric deep learning methods for small molecule drug discovery grounded in physics and chemistry. Specifically, I am using deep learning for quantum-level representations of molecules as a precursor for tasks in lead molecule optimization such as property prediction and protein-ligand binding affinity prediction.

Sam Money-Kyrle (DPhil)
Accurate prediction of molecular properties, such as protein-ligand affinity, off-target binding, toxicity, and mutagenicity, is an intrinsic component of small molecule drug discovery. My research focuses on the development of robust and generalisable computational tools that elucidate greater structural understanding of the interactions between small molecules and proteins. I am particularly interested in the application of novel deep learning methods and evaluating whether these approaches are capable of learning inherent biochemistry binding interactions.

Charlie Clark (DPhil)
The chemokine signalling network drives inflammation and therefore many inflammatory diseases. However, it has evolved to be highly redundant to resist pathogenic shutdown, and so successful anti-chemokine therapeutics must target multiple chemokines simultaneously. During my DPhil, I will apply experimental and computational techniques to this ‘poly-pharmacological’ problem by characterising promiscuous therapeutics that can target multiple chemokines simultaneously and tackle chemokine-driven inflammatory disease.

Sanaz Kazeminia (DPhil)
I focus on structure-based drug design (SBDD) which uses 3D protein structures to design small molecules in a pocket-aware manner. My interest is primarily on the application of diffusion models in this space, which gradually add noise from molecular structures in three-dimensional space to generate novel compounds that match target protein binding sites. The goal is to generate molecules with high binding affinities to their targets which are synthetically accessible and chemically sound. I am interested in exploring the application of constraints on these models, new architectures for complex protein-ligand systems and the integration of a large language model to make these tools accessible to medicinal chemists.

Nicholas Runcie (DPhil)
Numerous generative models have been developed to aid in the design of small molecule drugs. However, these models typically generate thousands of suggestions, leaving it unclear how to select which molecules to synthesize next. My research focuses on bridging the gap between generative models and chemists; I aim to enhance the usability and applicability of these models in drug discovery pipelines. To achieve this, I will explore hypothesis generation, hypothesis summarization, and model interpretability, combining my expertise in medicinal chemistry with machine learning techniques.

James Broster (DPhil)
I develop machine learning models to improve molecular docking by generating biologically meaningful and physically plausible poses that recover key molecular interactions. My work focuses on creating tools for real-world drug discovery applications, accounting for speed, accuracy, and the ability to account for protein flexibility.

Alvaro Prat (DPhil)
I am a first year PhD student working in both Computational Statistics and Machine Learning (CSML) and Oxford Protein Informatics (OPIG) groups. My current research interests pivot around developing robust and scalable generative models for structured data. I am also interested in low data regimes and advanced learning frameworks which focus on maximising information retrieval and signal propagation. I am excited to work alongside my supervisors, Yee Whye Teh (Oxford & DeepMind), Garrett Morris & Charlotte Deane (Oxford) to develop cutting edge solutions to enable accelerated & reliable drug discovery.